NatureScot Research Report 1328 - The ecological value of Sustainable Drainage Systems (SuDSs) in maintaining genetic diversity in urban areas
Year of publication: 2024
Authors: Robert Jehle (University of Salford), Jeanette Hall (NatureScot), Samantha A. Hook (University of Salford), Sarenta King (University of Salford), Kirsty MacArthur (Edinburgh Napier University), Alexandre Miró (Highland Amphibian and Reptile Project), Marcia Rae (NatureScot), David O'Brien (NatureScot)
Cite as: Jehle, R., Hall, J., Hook, S.A., King, S., MacArthur, K., Miró, A., Rae, M. and O'Brien, D. 2024. The ecological value of Sustainable Drainage Systems (SuDSs) in maintaining genetic diversity in urban areas. NatureScot Research Report 1328.
Keywords
amphibians; SuDS; microsatellites; genetic connectivity; urban ecology; land use policy; common frog; Rana temporaria
Background
Wildlife is increasingly residing in human-modified habitats such as urban spaces. While this predominately poses a threat to most taxa, it also offers opportunities in situations when natural habitats have become increasingly sparse. For the maintenance of within-species genetic variation, one of the three strands of biodiversity conservation, urban habitats can therefore harbour important population segments of given species.
The aim of the report was to use genetic markers to address the question whether common frogs (Rana temporaria) which breed in sustainable urban drainage system (SuDS) ponds suffer from an increased level of genetic erosion compared to populations in more rural areas. Studies in other small European cities have found low levels of diversity compared to the wider countryside and this has been associated with habitat fragmentation, due to roads and other artificial barriers. However, we hypothesised that the extensive network of green and blue infrastructure in Inverness might counteract isolation. To investigate this, levels of genetic variation and its temporal changes were compared between 22 urban and suburban SuDS ponds in Inverness with 12 counterpart populations from the city’s surroundings in 2015 and 2019.
Main findings
- In and around Inverness, changes in genetic variation over time were indiscernible between (sub)urban and rural common frog (Rana temporaria) populations.
- SuDS ponds harbour the same levels of genetic variation as rural ponds, with no evidence of inbreeding in any of the study populations
- The ecological quality of urban drainage systems was unrelated to levels of genetic variation.
- Genetic distances between populations were on average larger between (sub)urban populations than between rural populations.
Acknowledgements
We are grateful to the landowners for allowing access to their properties, and for their enthusiasm for the research.
1. Introduction
The assessment of the Intergovernmental Science-Policy Platform on Biodiversity and Ecosystem Services (‘IPBES’, Díaz et al. 2019) has highlighted global pressures on biodiversity and its current state. The report listed the following direct pressures in order of worldwide importance: (1) changes in land and sea use; (2) direct exploitation of organisms; (3) climate change; (4) pollution; and (5) invasive alien species and zoonoses. In many cases land use change is driven by urbanisation, which makes a further overarching indirect driver to biodiversity loss, people’s disconnection with nature, particularly pertinent (Díaz et al. 2019).
Despite having some of the least populated rural areas in Europe, overall 71% of people in Scotland live in densely populated urbanised parts of the country (Scotland’s Census 2011). The construction of Sustainable Drainage Systems (SuDS) in urbanising areas, which facilitate water runoff through surface infrastructure rather than below-ground engineering approaches, has been mandatory in Scotland since 2003, and led to the creation of ‘green-blue infrastructure’ in otherwise built-over land. SuDS have been shown to provide suitable habitats for biodiversity, including for breeding and feeding, although it is still largely unclear whether they also form part of habitat networks for interconnected, sustainable populations of inhabiting species (O’Brien, 2015; Rae et al., 2019).
Amphibians are well recognised as the most severely declining vertebrate taxon worldwide (e.g. Beebee & Griffiths 2005; Luedtke et al., 2023). Changes in land use, particularly habitat loss and degradation through urbanisation and agricultural intensification have long been documented as a major factor in their decline (e.g. Cooke & Ferguson, 1976; Cushman, 2006). While understanding existing pressures forms the basis for finding conservation solutions, evidence-based conservation of amphibians however remains a somewhat neglected field, and rather little is known about their long-term survival and evolutionary potential in artificial environments (Smith et al., 2018; Godet & Devictor 2018; Davies et al. 2018; Grant et al. 2019). One approach to monitor the effect of land-use change on amphibian populations is through genetic means, documenting how urbanisation and fragmentation alters the extent and spatial distribution of genetic variation at the landscape scale (Schmidt & Garroway, 2021; Hahs et al., 2023).
Due to their glacial history, the British Isles have a naturally rather depauperate amphibian fauna (Beebee and Griffiths 2000). In Scotland, road deaths, habitat loss and climate change are their main threats, with a dependence on small water bodies such as ponds leaving amphibian populations vulnerable to loss or damage to such habitats (Downie et al. 2019). The common frog (Rana temporaria) has possibly the widest ecological niche among the amphibians native to Scotland, which allows its persistence in all terrestrial ‘EUNIS habitat categories’ (see Davies. Moss and O’Hill, 2004 for a description of EUNIS) and reproduction in man-made sites such as garden and urban ponds (McInerny & Minting, 2016). Reproduction takes place in spring, when adults congregate in ponds at broadly even sex ratios to form local populations. Females deposit spawn clumps containing in the order of 1,000 - 2,000 eggs, from which tadpoles hatch and largely metamorphose in the same season (Cummins, 1996). The common frog was among the first amphibian species used to document the population genetic consequences of human-induced habitat fragmentation (Reh & Seitz, 1990; Hitchings & Beebee, 1997), and subsequently became a model species to study how specific landscape features impede or promote genetic connectivity and levels of genetic variation under pervasive anthropogenic influence (Johansson et al., 2005; 2006, Safner et al., 2011; Saarikivi et al., 2013; Lenhardt et al., 2017). In Scotland, the common frog is also among 26 other species chosen to represent genetic diversity for the purposes of reporting on progress against the CBD’s Aichi targets (Hollingsworth et al. 2020; O’Brien et al. 2022), further recognising the importance of the species as an indicator.
The city of Inverness in the Scottish Highlands is characterised by a rapid recent expansion, which over the last decades has resulted in the construction of an array of SuDS (O’Brien, 2015). Previous studies have demonstrated that the local SuDS ponds are inhabited by amphibian communities which are generally indiscernible from those of rural surrounding areas, despite marked variation in their ecological quality depending on their level of maintenance and design (O’Brien, 2015; Miró et al., 2018; Rae et al., 2019). The aim of this research is to expand on these studies by comparing the genetic structure of common frog populations inhabiting urban and suburban SuDS in Inverness with populations inhabiting rural ponds, and to determine whether genetic samples taken in two diffent years suggest that populations from SuDs ponds are characterised by genetic erosion. The present report parallels a recent journal article which also presents details of this study (Jehle et al. 2023).
2. Methods
2.1 Field sampling
A total of 34 populations were sampled in and around Inverness in April 2015, April 2019, or in both years (Table 1). The SuDS ponds that we considered (n = 22) represent a sub-sample of the sites described in Rae et al. (2019), and we used the Global Human Settlement Layer GHSL R2022A system (Pesaresi & Politis, 2022) to divide them into urban (n = 14; corresponding to the GHSL category Dense Urban Cluster) and suburban (n = 8; GHSL category Suburban) depending on their surrounding areas. The GHSL uses Copernicus Very High Resolution optical satellite imagery. Machine learning then identifies built-up surfaces at 2 m pixel resolution, and this is further subdivided into residential and non-residential. Land is then classified based on inhabitants per km2 and total settlement size. The rural ponds (n = 12; GHSL categories Low Density Rural and Very Low Density Rural combined) used in the study were situated up to a distance of about 20 km from Inverness, and are characterised in more detail in O’Brien et al. (2021a). The ecological quality of SuDS ponds was assessed by recording the presence or absence of 13 freshwater invertebrate groups with different pollution or eutrophication tolerance ranges following the OPAL protocol as described in Rae et al. (2019; see also Davies et al. 2011). Under the OPAL method, the surveyor assigns 10 points for less pollution tolerant animals (cased and caseless caddisflies, dragonflies, damselflies and alderflies), 5 points for moderately tolerant animals (mayflies, water beetles, water bugs, pond skaters and freshwater shrimps) and 1 point for tolerant animals (water slaters, snails and worms and worm-like animals), giving a maximum potential score of 78. Genetic sampling used a single embryo (egg) collected from separate clumps where possible, preserved in absolute ethanol until DNA extraction (Table 1). No precise information on adult population sizes, population age structures or growth and size of individuals is available.
Table 1. Details of sampling sites and descriptive population genetic parameters across 34 Rana temporaria populations in and around Inverness (Scotland). n: genetic sample sizes collected in two sampling years. Adapted from Jehle et al. (2023).
Urban
|
Sampling site |
National Grid Reference |
n (2015/2019) |
|---|---|---|
|
AA |
NH 67100 42136 |
13 (0/13) |
|
BA |
NH 69000 44010 |
10 (0/10) |
|
BB |
NH 69017 44011 |
16 (5/11) |
|
BD |
NH 71372 45055 |
15 (7/8) |
|
IP |
NH 68782 43278 |
14 (0/14) |
|
SD |
NH 64398 44565 |
11 (0/11) |
|
SP |
NH 67295 42107 |
27 (18/9) |
|
TA |
NH 71708 44979 |
10 (10/0) |
|
WC |
NH71519 45111 |
17 (8/9) |
|
WF |
NH 71820 44732 |
9 (9/0) |
|
WH |
NH 71904 44662 |
22 (10/12) |
|
WHR |
NH 66834 42423 |
22 (10/12) |
|
WP |
NH 71646 45212 |
8 (8/0) |
|
WO |
NH 68917 44160 |
14 (14/0) |
Suburban
|
Sampling site |
National Grid Reference |
n (2015/2019) |
|---|---|---|
|
BAA |
NH 70069 42351 |
10 (10/0) |
|
DV |
NH 67281 41847 |
10 (10/0) |
|
FA |
NH 67159 41965 |
10 (10/0) |
|
GR |
NH 69399 42517 |
10 (0/10) |
|
HFR |
NH 66477 41715 |
10 (10/0) |
|
HH |
NH 69068 45577 |
22 (9/13) |
|
IC |
NH 69221 45070 |
10 (10/0) |
|
MN |
NH 66968 41605 |
21 (10/11) |
Rural
|
Sampling site |
National Grid Reference |
n (2015/2019) |
|---|---|---|
|
AWH |
NH 59220 43830 |
21 (9/12) |
|
BW |
NH 47930 57240 |
9 (9/0) |
|
DC |
NH 63190 42000 |
11 (11/0) |
|
HP |
NH 59120 53680 |
21 (11/10) |
|
KM |
NH 60120 44180 |
27 (14/13) |
|
LL |
NH 53590 49720 |
9 (9/0) |
|
NSE |
NH 64320 42650 |
15 (5/10) |
|
NSW |
NH 64270 42540 |
21 (10/11) |
|
PH |
NH 86420 50310 |
20 (8/12) |
|
RO |
NH 6376044270 |
23 (8/15) |
|
SN |
NH 63894 44129 |
9 (0/9) |
|
TH |
NH 57620 54430 |
24 (11/13) |
2.2. Laboratory work
Samples were genotyped at the seven previously characterised common frog microsatellite loci BGF048, BGF053, BGF106, BGF142, BGF157, BGF250 and BGF258 (see Matsuba & Merilä, 2009); the specific loci were chosen due to their high number of alleles and the tri/tetranucleotide nature of repeat motifs for straightforward scoring. PCRs contained approximately 10 ng DNA, 5 pmol (5 mmol/L) of each primer, 0.15 mmol/L of each deoxynucleotide triphosphate (dNTP), 1.5 mmol/L MgCl2, and 0.5–1.0 U Taq polymerase (Advanced Biotechnologies, Columbia, MD) in the manufacturer’s buffer, at a total volume of 10 μl. The PCR profiles were 94 °C for 2 min, followed by 35 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s. Primers were labelled with fluorochromes (FAM, HEX or AT-550), and separated by capillary electrophoresis using an ABI 3130 Genetic Analyser (Applied Biosystems), either in-house at the University of Salford or commercially through Macrogen. Fragments were sized using Peak Scanner Software v1.0 (Applied Biosystems).
2.3 Data analysis
In a first set of analyses, we addressed the question of whether samples combined from the two sampling years 2015 and 2019 can be pooled for joint spatial analyses using the Bayesian clustering approach implemented in the software Structure 2.3.4 (originally described in Pritchard et al., 2000). The approach assigns individual genotypes to a predefined number of clusters (K) in a given sample (X), in order to achieve Hardy–Weinberg and linkage equilibrium. We estimated the ln posterior probabilities for K = 1 (implying one gene pool) or K = 2 (implying that the sampling years 2015 and 2019 represent different gene pools) for the 13 populations for which at least eight individuals were sampled in both study years (Table 1, n = 4, 2 and 7 for urban, suburban and rural populations, respectively), followed by calculating P (K|X) using Bayes’ rule. Results were obtained from 106 runs after 105 burn-ins, without allowing for admixture and using the correlated frequency model as implemented in the Structure software.
After showing that the site-specific samples are better characterised by assuming a single gene pool (see below), we computed observed (Ho) and expected (He) heterozygosities, departures from Hardy–Weinberg equilibria at each locus with Bonferroni corrections to give table-wide significance levels of P = 0.05, and measures of Fis for each population using Genepop on the Web, employing the implemented Markov Chain method (106 runs) to obtain unbiased estimates of Fisher’s exact tests (Rousset, 2008). FSTAT (Goudet, 1995) was used to obtain estimates of allelic richness based on the minimum population sample size. The spatial structure of populations was further investigated using the algorithm implemented in BAPS 6.0 (Cheng et al., 2013), to distinguish an enforced substructure (in our case, defined on the basis of ponds) from potentially more meaningful partitions reflected in the data set (for details see Corander et al., 2003). Bayesian posterior distributions are derived from an MCMC algorithm (we considered 500,000 runs after 100,000 burn-ins), and we set a lower probability bound of 0.05 for partitions to be considered in a final model. Patterns of pairwise spatial genetic differentiation between ponds were further assessed with Fst values also derived in Genepop, regressing Fst/(1- Fst) against geographic distance to test for scenarios of isolation-by-distance using Mantel tests as implemented in the software IBD (Bohanak, 2002). For the 13 populations for which at least eight individuals were sampled in both study years (Table 1, n = 4, 2 and 7 for urban, suburban and rural populations, respectively), we also investigated whether the two temporal samples differed in their overall levels of genetic variation. Based on published information on the common frog (Miaud et al., 1999), the four-year interval between sampling years broadly represents one generational turnover.
3. Results
This report considers a total of 521 genotypes across the studied 34 populations (n = 208, 103 and 210 for urban, suburban and rural populations, respectively). These genotypes encompassed 273 and 248 samples collected in 2015 and 2019, respectively, resulting in an average of 14.2 total samples per population (range 8-27). The overall PCR success rate across all loci was 83.0%, with a minimum per-population sample size per locus of n = 3 for the calculation of allelic richness values.
According to the algorithm implemented in the software Structure, posterior probabilities that samples from a given pond represent a single genetic cluster (K = 1) ranged from 0.77 (rural population PH) to 1.00 (rural population AWH and suburban population MN) with a median of 0.97, confirming that the two sampling years can be merged for joint spatial analyses (Table 2). The mean number of alleles per locus ranged between 4.29 (rural population IP) and 10.14 (urban population WH), with an average of 6.70, 6.68 and 6.94 for urban, suburban and rural populations, respectively, and no significant differences between the groups (one-way ANOVA, d.f. = 2, F = 2.031, p = 0.15; Table 2). Corresponding mean values of allelic richness ranged from 3.53 (rural) over 3.67 (urban) to 3.89 (suburban), again without significant differences between the groups (d.f. = 2, F = 0.08, p = 0.92; Figure 1). In total, 31 out of 34 populations (91.2%) were in Hardy-Weinberg equilibrium at a Bonferroni-adjusted p value of 0.0015, with heterozygosities exceeding expected values in 22/34 (64.7%) of cases (Table 2). High heterozygosities were also reflected in slightly negative mean Fis values (urban: -0.04; suburban: -0.03; rural: -0.05; Figure 1). For the urban and suburban populations combined, there was no significant correlation between OPAL scores and the average number of alleles (Spearman rank correlation: rs = -0.19, p = 0.53) or allelic richness (rs = -0.16, p = 0.60).
Table 2. Descriptive population genetic parameters across 34 Rana temporaria populations in and around Inverness (Scotland). p (K=1): probability that the two sampling years represent a single gene pool (for more details see text); Ho: observed mean heterozygosity; He: expected mean heterozygosity, where * denote deviations from population-wide Hardy-Weinberg equilibria at a Bonferroni-corrected p value of 0.0015; AL: mean number of alleles per locus; PA: number of private alleles. Expanded from Jehle et al. (2023).
Urban
|
Sampling site |
p (K=1) |
Ho |
He |
AL |
PA |
|---|---|---|---|---|---|
|
AA |
- |
0.68 |
0.77 |
5.57 |
0 |
|
BA |
- |
0.95 |
0.81 |
5.86 |
0 |
|
BB |
- |
0.77 |
0.75 |
6.57 |
1 |
|
BD |
- |
0.78 |
0.74 |
6.29 |
0 |
|
IP |
- |
0.56 |
0.57 |
4.29 |
0 |
|
SD |
- |
0.63 |
0.64 |
4.33 |
0 |
|
SP |
- |
0.77 |
0.81 |
9.86 |
0 |
|
TA |
- |
0.82 |
0.78 |
6.14 |
0 |
|
WC |
- |
0.67 |
0.70 |
6.43 |
0 |
|
WF |
- |
0.91 |
0.78 |
5.57 |
1 |
|
WH |
- |
0.79 |
0.81 |
10.14 |
2 |
|
WHR |
- |
0.78 |
0.81 |
8.43 |
0 |
|
WP |
- |
0.84 |
0.75 |
7.00 |
0 |
|
WO |
- |
0.94 |
0.81* |
7.29 |
0 |
Suburban
|
Sampling site |
p (K=1) |
Ho |
He |
AL |
PA |
|---|---|---|---|---|---|
|
BAA |
- |
0.69 |
0.76 |
5.57 |
1 |
|
DV |
- |
0.91 |
0.81 |
6.57 |
0 |
|
FA |
- |
0.89 |
0.81 |
6.57 |
0 |
|
GR |
- |
0.71 |
0.68 |
4.29 |
1 |
|
HFR |
- |
0.84 |
0.82 |
7.00 |
1 |
|
HH |
- |
0.75 |
0.80* |
9.57 |
2 |
|
IC |
- |
0.93 |
0.83 |
7.14 |
0 |
|
MN |
- |
0.82 |
0.81* |
6.71 |
1 |
Rural
|
Sampling site |
p (K=1) |
Ho |
He |
AL |
PA |
|---|---|---|---|---|---|
|
AWH |
- |
0.78 |
0.77 |
6.14 |
0 |
|
BW |
- |
0.74 |
0.68 |
5.40 |
0 |
|
DC |
- |
0.77 |
0.73 |
4.57 |
1 |
|
HP |
- |
0.79 |
0.76 |
7.57 |
2 |
|
KM |
- |
0.80 |
0.77 |
10.14 |
1 |
|
LL |
- |
0.90 |
0.79 |
5.43 |
0 |
|
NSE |
- |
0.78 |
0.80 |
7.43 |
3 |
|
NSW |
- |
0.76 |
0.74 |
7.29 |
0 |
|
PH |
- |
0.74 |
0.75 |
6.86 |
0 |
|
RO |
- |
0.70 |
0.71 |
8.43 |
1 |
|
SN |
- |
0.75 |
0.66 |
4.43 |
0 |
|
TH |
- |
0.84 |
0.77 |
9.57 |
0 |
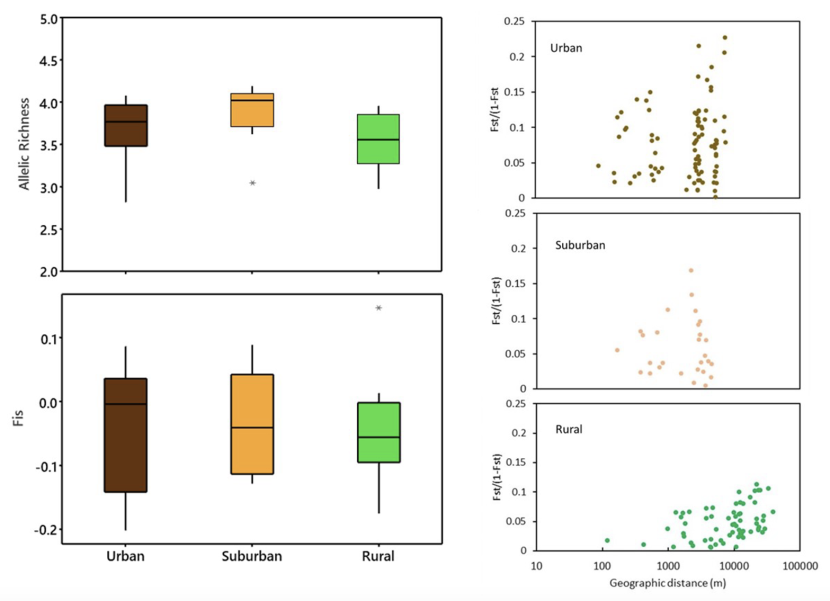
Left panel: allelic richness and Fis values; right panel: pairwise comparisons between log-transformed geographic and genetic distances, with a significant correlation for rural populations only (for details see text). Adapted from Jehle et al. (2023).
The graph shows there is no statistically significant difference between the allelic richness for each with mean values ranging from 3.53 in rural, though 3.67 for urban to 3.89 for suburban populations. Similarly the figure shows little difference in Fis , a measure of inbreeding. Urban, suburban and rural populations all show negative Fis values which suggests that inbreeding is not an issue. Mean urban Fis is -0.04; suburban: -0.03; and rural: -0.05. The figure also has a panel showing pairwise comparisons between log-transformed geographic and genetic distances. This shows no isolation by distance for urban or suburban populations but highly significant isolation by distance for rural populations.
Across all considered ponds, private alleles were least common in urban populations (n = 4), followed by suburban (n = 6) and rural populations (n = 8). Average pairwise Fst values were larger between urban (0.07) and suburban (0.05) than between rural (0.04) populations, suggesting an overall stronger partition of genetic variation in built-over areas despite higher overall geographic proximity. This is further reflected by isolation-by-distance scenarios which are absent in suburban (Z = 5.27, p = 0.54) and urban populations (Z = 24.40, p = 0.60), and highly significant for rural populations (Z = 12.55, p > 0.01, Figure 1). The algorithm as implemented in the software BAPS reduced the 34 ponds to five genetic clusters (Figure 2). One cluster consisted of the single, spatially isolated urban SuDS pond IP, and a further cluster consisted of the spatially adjacent but differentially classified ponds SN (rural) and SD (urban). The remaining three clusters comprised six, seven and 18 populations each (Figure 2).
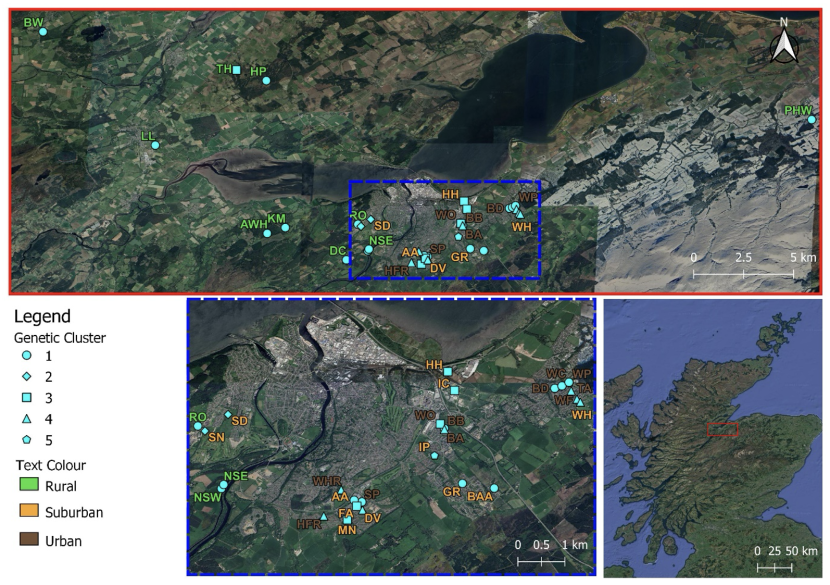
The classification of Rana temporaria study ponds is according to the Global Human Settlement Layer system (rural, suburban or urban) and their partition into five genetic clusters as identified using the software BAPS. Sampling site designations are identical to those shown in Table 1. For more details see text. Adapted from Jehle et al. (2023).
The 34 study ponds are coloured to show their classification according to the Global Human Settlement Layer system (rural, suburban or urban) and into five genetic clusters. Cluster 2 is made up of two suburban ponds in western Innerness. Cluster 3 includes three urban and suburban ponds in northern Inverness which are connected by green infrastructure, two closely connected suburban ponds in southern Inverness and one rural pond about 15km north west of Inverness. Cluster 4 is made up of urban and suburban ponds in eastern and southern Inverness and Cluster 5 is a single spatially isolated pond in south-east Inverness. The remaining sites are part of Cluster 1 which includes most of the rural ponds both east and west of Inverness, as well as two suburban ponds in south-eastern Inverness and urban ponds in east and south Inverness.
The comparison of levels of genetic variation between the two sampling years 2015 and 2019 revealed no marked differences between rural, urban and suburban sites, with the majority of populations across all sites however characterised by a slight decrease in allelic richness (Figure 3).
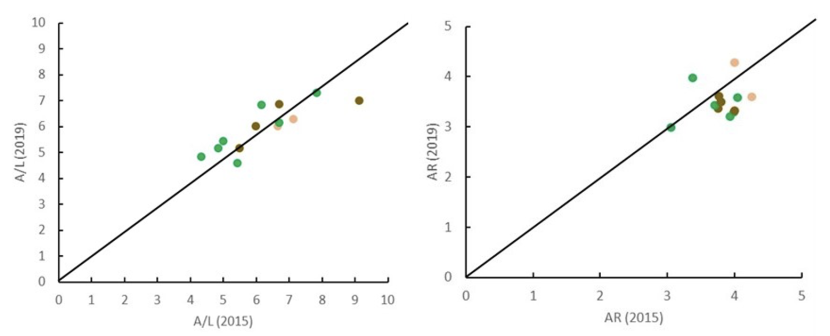
Values above/below the diagonal represent increases/decreases over time. Green symbols: rural populations; orange symbols: suburban populations; brown symbols: urban populations. The graphs show little change over the study period, with a slight decline in allelic richness and no differences between urban, suburban and rural sites. This suggests that urban sites are no more prone to loss of genetic diversity than rural ones. Adapted from Jehle et al. (2023).
The first graph compares alleles per locus in 2015 and 2019 for each population. The second compares allelic richness for each population in 2015 and 2019. Values above the line in the graph show increases over time, those below the line show decreases. The graph shows a slight increase in alleles per locus and a slight decrease in allelic richness over time. There was no marked difference between rural suburban and urban sites. All of the populations are close to the line in both graphs with the exception of one urban population which shows a more sizeable decrease in alleles per locus.
4. Discussion
Although the Convention on Biological Diversity includes species, ecosystem and genetic conservation within its remit (CBD 1992), genetic conservation sometimes receives less attention than species or ecosystem conservation (Pérez-Espona and ConGRESS Consortium, 2017; Hoban et al. 2020; Hoban et al., 2023). This report on the common frog considers all three levels of biodiversity, comparing genetic variation as a measure of evolutionary potential between different habitats and thus contributing to species management plans. It also adds to our knowledge of one of the species selected for Scotland’s Genetic Diversity Scorecard (Hollingsworth et al. 2020). We therefore hope that the findings of this report will contribute to both national biodiversity management policies as well as local land use planning.
The present study builds upon existing work which demonstrates that the rather recently constructed SuDS ponds in Inverness provide suitable breeding habitats for the locally occurring amphibian assemblage (O’Brien, 2015; Rae et al., 2019). In rural areas of Scotland as a whole, on the other hand, changes in agricultural practice in the 20th century have led to heavy losses of small aquatic sites such as freshwater ponds through infilling, pollution and natural succession (Walton et al. 2019). Given that urban green-blue spaces such as SuDS therefore have the potential to serve as important habitats, we used the common frog as an example species to investigate whether urban settings have compromise the evolutionary potential of residing populations. We found that suburban and urban SuDS ponds, despite overall increasing local levels of genetic differentiation, are not characterised by higher levels of genetic erosion compared to rural sites. Our case study adds to the growing evidence that generalisations on the effects of human-induced habitat fragmentation on amphibians are difficult to infer (e.g. Schmidt & Garroway, 2021). It also reinforces that urban habitats in cities such as Inverness can provide an important contribution to the preservation of within-species genetic diversity for those species which can persist in human-modified areas.
Given that our study site in Inverness is situated in the north-western periphery of the common frog’s western Palearctic range, the overall high levels of allelic diversity revealed by our study is noteworthy. It also exceeds those found in another microsatellite-based study conducted in Scotland across favourable habitat in the absence of interpopulation barriers, such as busy roads (Muir et al., 2013). Although it is impossible to discard local, population-specific reasons for this observation, our choice of loci from a pool of 145 candidate markers offered by Matsuba & Merilä (2009) was based on the most polymorphic loci available and may in part explain this difference. We also encountered observed heterozygosities which for the majority of populations were above their expected values under Hardy-Weinberg equilibrium (Table 2). While positive relationships between genetic diversity and fitness generally apply to rather harsh conditions such as species’ range edges and have been documented in the common frog before (Hitchings & Beebee, 1997; Lesbarrères et al., 2005), we rather attribute this observation to our sampling regime which avoided the random collection of full siblings by only analysing one egg per clump. High levels of observed heterozygosities also further support that the pooling of individuals from two sampling years for spatial inferences was justified, as an unconsidered substructure would reduce Ho to below He through the Wahlund effect (Wahlund, 1928).
A main finding arising from the present report is that urban and suburban SuDS ponds harbour the same levels of genetic variation as rural ponds, with no evidence of inbreeding in any of the study populations. Our genetic data suggest that gene flow might be responsible for counteracting the negative consequences of drift, and is in line with the findings from the clustering approach which revealed a wide partitioning of 34 ponds into five genetic groups whereby SuDS ponds are not separated from their rural counterparts. Our findings, however, contrast with a previous study of British urban common frog populations in Brighton (Hitchings & Beebee, 1997) which revealed reduced levels of genetic variation combined with inbreeding depression in urban populations. That study predated widespread adoption of SuDS in the UK and none were included. The city of Inverness has undergone a particular rapid growth since the late 20th century, resulting in an urban area expansion which led to the rather recent creation and modification of the SuDS ponds under study (O’Brien, 2015). This suggests two possible explanations. Either there has not yet been time for genetic erosion to occur, or the green infrastructure associated with SuDSs provides both suitable breeding habitat and functional connectivity of terrestrial habitat favourable to conservation of genetic variation in this species. The demographic consequences of habitat modification and population isolation might therefore still accumulate over time. However, since several of the ponds sampled have existed for over 20 years (approximately five generations for the common frog, see Miaud et al. 2000), signs of genetic erosion would be expected if isolation effects were strong.
Facilitated by a high plasticity in breeding behaviour, diet and larval development, the common frog possesses a wide ecological niche which is known to include human-influenced habitats (Johansson et al., 2005; McInerny & Minting, 2016). It is therefore not overly surprising that we found no clear link between pond habitat quality as measured by OPAL and genetic variation. SuDS ponds in Inverness have been found to have higher OPAL scores compared to Scotland and Britain as a whole, confirming their high value as breeding habitat for our study species (O’Brien et al., 2015, Rae et al., 2019). Environmental conditions are however known to influence vital demographic parameters such as individual longevity and growth in R. temporaria populations (e.g. Miaud et al., 1999; Richter-Boix et al., 2010). From a conservation perspective, it would therefore be beneficial to compare levels of recruitment, generational turnover and habitat-dependent life-history parameters such as diet between SuDS ponds and rural sites in future studies.
The distribution of genetic variation in given landscapes arises from a combination of spatial and temporal processes. Ponds from rural areas were characterised by a scenario of isolation-by-distance which represents levels of connectivity that are proportional to geographic proximity. This matches previous studies which noted low levels of artificial barriers to amphibian movement such as major roads and human settlements in the study area as a whole (O’Brien et al., 2021a; b). Isolation-by-distance was however absent in urban and suburban ponds, whose genetic makeups appeared to be dominated by drift, or gene flow which is uncoupled from geographic distance. This may be related to the distribution of SuDS in green ‘fingers’ of land within the city which means that two ponds may be geographically close to each other but separated by unfavourable habitat. While landscape genetic analyses are beyond the scope of the present work, it is noteworthy that we revealed overall higher levels of pairwise genetic differentiation (Fst values) between SuDS ponds compared to rural sites, despite a higher overall number of population-private alleles in the latter. Aside from modified or reduced patterns of gene flow, the observed breakdown of isolation-by-distance in urban and suburban areas might therefore also be linked to the spatial scale of investigation: rural populations covered a wider area than their urban and suburban counterparts, which might have led to a more pronounced genetic signal of spatial differentiation.
In our temporal comparison of samples collected within a 4-year interval, we revealed no tendency for changes in mean numbers of alleles per locus, alongside a tendency for a reduction of allelic richness across (sub)urban as well as rural sites. This seems likely to be linked to the rather limited per-population, per-locus sample size rather than to a scenario of genetic drift, which would result in a higher probability of rare alleles becoming lost than common alleles. Significantly for our study, this supports the evidence that genetic erosion is not significant in our urban populations. Our inferences nevertheless reinforce the general importance of temporal genetic monitoring of populations. We however refrained from, for example, using the temporal samples to compare genetic effective population sizes between urban, suburban and rural populations, due to the expected large confidence intervals under the given sampling regime and the general caution which is recommended for such an approach due to the amphibian life history (see e.g. Kimble et al., 2023). Given the increased availability of amphibian genetic information and tools, such as genomes and transcriptomes (for R. temporaria see Price et al., 2015; Streicher et al., 2021), future research could focus on how habitats such as SuDS ponds shape the distribution of adaptive genetic variation, in response to mounting evidence of selection for distinctive phenotypic traits such as reduced mobility, larger body size and fewer offspring in dense urban settings (see also Hahs et al., 2023).
What do our results tell us for the maintenance of within-species genetic variation of amphibians such as the common frog across human-modified landscapes in Scotland? When suitably managed, SuDS ponds appear to provide nature-friendly neighbourhoods which enable the retention of evolutionary potential for common frog populations. This is an encouraging finding, given that the density of such sites in built-over areas can be higher than for example in agricultural land which has suffered a marked loss of amphibian breeding ponds in recent decades (Hill et al., 2007; Walton et al., 2019). From a policy perspective, attempts to maximise biodiversity conservation opportunities require an understanding of how different habitats contribute to the evolutionary potential of given species. Our findings reinforce the idea that well-designed and managed urban ecosystems can harbour an integral share of the overall genetic diversity for those species that are able to use them for reproduction. While not the case for amphibians in our study area, urban environments can also act as filters which reduce the overall number of species (Hamer & McDonnell, 2008; Hamer & Parris, 2011). Our findings support the emphasis on increasing the quality and connectivity of blue-green infrastructure both locally, for example in the developing Scottish Biodiversity Strategy (Scottish Biodiversity Strategy, 2023) and globally through Target 12 of the Convention on Biological Diversity (CBD, 2023). They also add an often-missing genetic dimension to our understanding of the importance of connectivity (Hoban et al., 2020) and contribute to the development of an evidence-based approach to conservation. Whilst the lifecycles of other Scottish amphibians may be different to that of the common frog, this set of SuDSs has been studied over 12 years and our work thus also highlights the importance of long-term studies to inform conservation interventions.
References
Beebee, T.J. and Griffiths, R.A. 2005. The amphibian decline crisis: a watershed for conservation biology? Biological Conservation, 125, 271–285.
Bohonak, A.J. 2002. IBD (Isolation by Distance): a program for analysis of isolation by distance. Journal of Heredity, 93, 153–154.
Cheng, L., Connor, T.R., Sirén, J., Aanensen, D.M. and Corander, J. 2013. Hierarchical and spatially explicit clustering of DNA sequences with BAPS software. Molecular Biology and Evolution, 30, 1224–1228.
Cooke, A.S. and Ferguson, P.F. 1976. Changes in status of the frog (Rana temporaria) and the toad (Bufo bufo) on part of the East Anglian Fenland in Britain. Biological Conservation, 9, 191–198.
Corander, J., Waldmann, P. and Sillanpää, M.J. 2003. Bayesian analysis of genetic differentiation between populations. Genetics, 163, 367–374.
Cummins, C.P. 1996. Temporal and spatial variation in egg size and fecundity in Rana temporaria. Journal of Animal Ecology, 55, 303–316.
Cushman, S.A. 2006. Effects of habitat loss and fragmentation on amphibians: a review and prospectus. Biological Conservation, 128, 231–240.
Davies, C.E., Moss, D. and O'Hill, M. 2004. EUNIS habitat classification. Revision 2004. European Environment Agency. European Topic Centre on Nature Protection and Biodiversity.
Davies, L., Bell, J.N.B., Bone, J., Head, M., Hill, L., Howard, C., Hobbs, S.J., Jones, D.T., Power, S.A., Rose, N., Ryder, C., Seed, L., Stevens, G., Toumi, R., Voulvoulis, N. and White, P.C.L. 2011. Open Air Laboratories (OPAL): A community-driven research programme. Environmental Pollution, 159, 2203–2210.
Davies, T., Cowley, A., Bennie, J., Leyshon, C., Inger, R., Carter, H., Robinson, B., Duffy, J., Casalegno, S., Lambert, G. and Gaston, K., 2018. Popular interest in vertebrates does not reflect extinction risk and is associated with bias in conservation investment. PloS One, 13, e0203694.
Díaz, S. et al. 2019. Pervasive human-driven decline of life on Earth points to the need for transformative change. Science, 366, eaax3100.
Downie, J.R., Larcombe, V. and Stead, J. 2019. Amphibian conservation in Scotland: A review of threats and opportunities. Aquatic Conservation: Marine and Freshwater Ecosystems, 29, 647-654.
Godet, L. and Devictor, V. 2018. What conservation does. Trends in Ecology & Evolution, 33, 720-730.
Goudet, J. 1995. FSTAT (Version 1.2): a computer program to calculate F-statistics. Journal of Heredity, 86, 485-486.
Grant, E.H.C., Muths, E., Schmidt, B.R. and Petrovan, S.O. 2019. Amphibian conservation in the Anthropocene. Biological Conservation, 236, 543–54.
Hahs, A.K. et al. 2023. Urbanisation generates multiple trait syndromes for terrestrial animal taxa worldwide. Nature communications, 14(1), 4751.
Hamer, A.J. and McDonnell, M.J. 2008. Amphibian ecology and conservation in the urbanising world: A review. Biological Conservation, 141, 2432–2449.
Hamer, A.J. and Parris, K.M. 2011. Local and landscape determinants of amphibian communities in urban ponds. Ecological Applications, 21, 378–390.
Hill, M.J., Biggs, J., Thornhill, I., Briers, R.A., Gledhill, D.G., White, J.C., Wood, P.J. and Hassall, C. 2017. Urban ponds as an aquatic biodiversity resource in modified landscapes. Global Change Biology, 23, 986–999.
Hitchings, S.P. and Beebee, T.J.C. 1997. Genetic substructuring as a result of barriers to gene flow in urban Rana temporaria (common frog) populations: implications for biodiversity conservation. Heredity, 79, 117–127.
Hoban, S. et al. 2021. Global commitments to conserving and monitoring genetic diversity are now necessary and feasible. BioScience, 71, 964–976.
Hoban, S. et al. 2023. Genetic diversity goals and targets have improved, but remain insufficient for clear implementation of the post-2020 global biodiversity framework. Conservation genetics, 24(2), 181–191.
Jehle, R., Hall, J., Hook, S.A., King, S., MacArthur, K., Miró, A., Rae, M. and O’Brien, D. 2023. High evolutionary potential maintained in common frog (Rana temporaria) populations inhabiting urban drainage ponds. Diversity, 15(6), p.738.
Johansson, M., Primmer, C.R. and Merilä, J. 2006. History vs. current demography: explaining the genetic population structure of the common frog (Rana temporaria). Molecular Ecology, 15, 975-983.
Johansson, M., Primmer, C.R., Sahlsten, J. and Merilä, J. 2005. The influence of landscape structure on occurrence, abundance and genetic diversity of the common frog, Rana temporaria. Global Change Biology, 11, 1664–1679.
Kimble, S.J.A., Unger, S.D. and Williams, R.N. 2023. Genetically derived effective population size estimates of herpetofaunal species should be used with caution. Journal of Wildlife Management, 87, e22340.
Lenhardt, P.P., Brühl, C.A., Leeb, C. and Theissinger, K. 2017. Amphibian population genetics in agricultural landscapes: does viniculture drive the population structuring of the European common frog (Rana temporaria)? PeerJ, 5, e3520.
Lesbarrères, D., Primmer, C.R., Laurila, A. and Merilä J. 2005. Environmental and population dependency of genetic variability‐fitness correlations in Rana temporaria. Molecular Ecology, 14, 311–323.
Luedtke, J.A. et al. 2023. Ongoing declines for the world’s amphibians in the face of emerging threats. Nature, 622, 308–314.
Matsuba, C. and Merilä, J. 2009. Isolation and characterization of 145 polymorphic microsatellite loci for the common frog (Rana temporaria). Molecular Ecology Resources, 9, 555–562.
McInerny, C. and Minting, P. 2016. The Amphibians & Reptiles of Scotland. Glasgow: The Glasgow Natural History Society.
Miaud, C., Guyétant, R. and Elmberg J. 1999. Variations in life-history traits in the common frog Rana temporaria (Amphibia: Anura): a literature review and new data from the French Alps. Journal of Zoology, 249, 61–73.
Miró, A., Hall, J., Rae, M. and O’Brien, D. 2018. Links between ecological and human wealth in drainage ponds in a fast-expanding city, and proposals for design and management. Landscape and Urban Planning, 180, 93–102.
Muir, A.P., Thomas, R., Biek, R. and Mable, B.K. 2013. Using genetic variation to infer associations with climate in the common frog, Rana temporaria. Molecular Ecology, 22, 3737–3751.
O’Brien, C.D. 2015. Sustainable drainage system (SuDS) ponds in Inverness, UK and the favourable conservation status of amphibians. Urban Ecosystems, 18, 321–331.
O’Brien C.D., Hall J.E., Miró A., O’Brien K., Falaschi M. and Jehle R. 2021a. Reversing a downward trend in threatened peripheral amphibian (Triturus cristatus) populations through interventions combining species, habitat and genetic information. Journal for Nature Conservation, 64, 126007.
O’Brien, C.D., Hall, J.E., Miró, A., O’Brien, K. and Jehle, R. 2021b. A co-development approach to conservation leads to informed habitat design and rapid establishment of amphibian communities. Ecological Solutions and Evidence, 2, e12038.
O'Brien, D. et al. 2022. Bringing together approaches to reporting on within species genetic diversity. Journal of Applied Ecology, 59(9), pp.2227-2233.
Pesaresi, M. and Politis P. 2022. GHS built-up surface grid, derived from Sentinel2 composite and Landsat, multitemporal (1975-2030). European Commission Joint Research Centre (JRC).
Price, S.J., Garner, T.W.J., Balloux, F., Ruis, C., Paszkiewicz, K.H., Moore, K. and Griffiths, A.G.F. 2015. A de novo assembly of the common frog (Rana temporaria) transcriptome and comparison of transcription following exposure to Ranavirus and Batrachochytrium dendrobatidis. PLoS ONE, 10, e0130500.
Pritchard, J. K., Stephens, M. and Donnelly, P. 2000. Inference of population structure using multilocus genotype data. Genetics, 155, 945–959.
Rae, M., Miró, A., Hall, J., O’Brien K. and O’Brien, D. 2019. Evaluating the validity of a simple citizen science index for assessing the ecological status of urban drainage ponds. Ecological Indicators, 98, 1–8.
Reh, W. and Seitz, A. 1990. The influence of land use on the genetic structure of populations of the common frog Rana temporaria. Biological Conservation, 54, 239–249.
Richter-Boix, A., Teplitsky, C., Rogell, B. and Laurila, A. 2010. Local selection modifies phenotypic divergence among Rana temporaria populations in the presence of gene flow. Molecular Ecology, 19, 716–731.
Rousset, F. 2008. Genepop’007: a complete reimplementation of the Genepop software for Windows and Linux. Molecular Ecology Resources, 8, 103–106.
Saarikivi, J., Knopp, T., Granroth A. and Merilä, J. 2013. The role of golf courses in maintaining genetic connectivity between common frog (Rana temporaria) populations in an urban setting. Conservation Genetics, 14, 1057–1064.
Safner, T., Miaud, C., Gaggiotti, O., Decout, S., Rioux, D., Zundel, S. and Manel, S. 2011. Combining demography and genetic analysis to assess the population structure of an amphibian in a human-dominated landscape. Conservation Genetics, 12, 161–173.
Sagvik, J., Uller, T. and Olsson, M. 2005. Outbreeding depression in the common frog, Rana temporaria. Conservation Genetics, 6, 205–211.
Schmidt, C. and Garroway, C.J. 2021. The population genetics of urban and rural amphibians in North America. Molecular Ecology, 30, 3918–3929.
Schmidt, C., Munshi-South, J., Dray, S. and Garroway, C.J. 2022. Determinants of genetic diversity and species richness of North American amphibians. Journal of Biogeography, 49, 2005–2015.
Scotland’s Census, n.d. Scotland’s Census 2011.
Scottish Government, 2022. Scottish Biodiversity Strategy to 2045: Tackling the Nature Emergency in Scotland.
Smith, T., Beagley, L., Bull, J., Milner‐Gulland, E.J., Smith, M., Vorhies, F. and Addison, P.F., 2020. Biodiversity means business: Reframing global biodiversity goals for the private sector. Conservation Letters, 13, e12690.
Streicher J.W. and Wellcome Sanger Institute Tree of Life Programme, 2021. The genome sequence of the common frog, Rana temporaria Linnaeus 1758, Wellcome Open Research, 6, 286.
United Nations, n.d. Convention on Biological Diversity.
Wahlund S. 1928. Zusammensetzung von Populationen und Korrelationerscheinungen vom Standpunkt der Vererbungslehre aus betrachtet. Hereditas, 11, 65–106.
Walton, P. et al. 2019. The State of Nature Scotland 2019. The State of Nature partnership.





